About Us:
Functional Imaging Group
The Functional Imaging Group in the Department of Radiation Oncology at the University of Michigan was established in 2003.
Overall goals of our group are to develop, validate and translate MR imaging technologies, including quantitative and automated models and methods for functional and metabolic imaging, tumor delineation, and rapid imaging to guide physiological adaptive radiation therapy of cancers. Currently, we are focusing on four major areas listed below.
Our research projects have been supported by NIH grants (4P01CA59827 (Ten Haken), 1U01CA183848(Cao/Eisbruch), RO1 NS064973(Cao), RO1 CA132834 (Cao), and RO1 EB016079 (Balter/Cao), industrial grant (Siemens), and our institute and department funds.
Project:
AI Based Tools for Detection and Delineation of Intracranial Metastases from MRI
AI based tools for detection and delineation of intracranial metastases from MRI
Academia-Industrial R01 (NIH CA262182-01): 71/2021-6/20/2026 (the 4 percentile score).
This is a collaborative project involved in investigators from medical physicists, radiologists, radiation oncologists and AI specialists from three academic institutes (UMich. NYU, UNC) and one industrial partner (AI team of Siemens Medical Solutions USA, Inc).
The goal of the project is to develop an AI based tool for automated detection and delienation of intracranial metastases from clinical diagnosis 3D MRI. An AI system that is optimized for accurate detection and delimitation of brain metastases from 3D MRI could have a large scale impact on brain metastasis management including diagnosis, treatment, and longitudinal therapy response assessment.
The database will contain more 1800 patients with approximately 2800 metastatic lesions from three academic clinics.
PI: Yue Cao, Ph.D., FAAPM
- University of Michigan Team: Yue Cao, Ph.D., James Balter, Ph.D., Michelle Kim, MD, Hemant Parmar, MD, Mohannad Ibrahim, MD, Yilun Sun, Ph.D.
- New York University Team: Hesheng Wang, Ph.D., Douglas Kondziolka, MD, Rajan Jain, MD
- University of North Carolina Team: Tong Zhu, Ph.D., Colette Shen, MD, Yueh Lee, MD
- Siemens Medical Solutions USA, Inc Team: Eli, Gibson , Ph..D., Youngjin Yoo, Ph.D., Thomas J. Re, MD
Select Publications
- R. Farjam, H. A. Parmar, D. C. Noll, C. Tsien, and Y. Cao. A New Approach for Computer-Aided Detection and Segmentation of Brain Metastases in Post-Gd T1-weighted MRI. Magnetic Resonance Imaging 30(6):824-36, 2012. PMID: 22521993. PMCID: PMC3932529
- F.C. Ghesu, B. Georgescu, A. Mansoor, Y. Yoo, E. Gibson, R.S. Vishwasnath, A. Balachandran, M. K. Kalra, R. Singh, S. R. Digumarthy, J. M. Balter, Y. Cao, S. Grbic, D. Comaniciu. Quantifying and leveraging predictive uncertainty for medical image assessment. Medical Image Analysis, Oct 13, 2020, 68:101855. DOI: 10.1016/j.media.2020.101855 PMID: 33260116
- Y. Yoo, T.J.Re, S.Liu, P. Ceccaldi, Y. Cao, J. M.Balter, E. Gibson. Evaluating deep learning methods in detecting and segmenting different sizes of brain metastases on 3D MRI. JMI (revision) 2021.
Deep Learning for MR Image Reconstruction and Physiological Parametric Map Estimation
Supported by RO1 EB016079 (James Balter/Yue Cao). The project is led by Yue Cao, Ph.D., FAAPM.
The goal of this project is to develop and test deep learning techniques for joint optimization of spatial and temporal sparsity sampling patterns in k-space and image reconstruction of dynamic and non-dynamic MRI and physiological parametric maps. Spatial and temporal sparsity in acquired data in MR imaging can be optimized for improving acquisition speed dramatically and achieving high quality images. Also, deep learning techniques can estimate physiological parameters with suboptimal data more accurately than conventional least-squares fitting of a pharmacokinetic model.
Select Publications
- Jiaren Zou, and Yue Cao. Joint Optimization of k-t Sampling Pattern and Reconstruction of DCE MRI for Pharmacokinetic Parameter Estimation. IEEE Transactions on Medical Imaging, 2022 Jun17. DOI: 10.1109/TMI.2022.3184261 10.1109/TMI.2022.3184261.
- Josiah Simeth, and Yue Cao. GAN and Dual-input two-compartment model based training of a neural network for robust quantification of contrast uptake rate in Gadoxetic acid-enhanced MRI. Medical Physics, 47(4):307-316, 2020. https://doi.org/10.1002/mp.14055
- J. Zou, J. M. Balter, Y. Cao. Estimation of pharmacokinetic Parameters from DCE-MRI by extracting long and short time-dependent features using an LSTM network. Medical Physics, 47(8):3447-3457, 2020.doi:10.1002/mp.14222.
- V. Agarwal, J. M. Balter, Y. Cao. Cascaded Attention Residual Non-Local Network (CARNL-Net): Image Reconstruction for Accelerated MR Imaging. Submitted to MRM 2021.
- J. Zou , J. Balter, and Y. Cao. Estimation of Pharmacokinetic Parameters from DCE MRI Using an Attention Bidirectional Long Short-term Memory Neural Network. International Journal of Radiation Oncology, Biology, Physics, 108(3): S130, 2020.
- J. Zou, J. Balter, and Y. Cao. Estimation of Pharmacokinetic Parameters from DCE-MRI by Extracting Long and Short Time-dependent Features Using a LSTM network. Proceedings of ISMRM 2020.
- Vihang Agarwal, James Balter, Yue Cao. Accelerating MR image acquisition using Cascaded Attention Residual U-Net (CARU-Net). AAPM, 2020.
- J. Zou, and Y. Cao. Joint Deep Learning-based Optimization of Subsampling Pattern and Reconstruction for Dynamic Contrast-Enhanced MRI. Proceedings of ISMRM 2021.
- Vihang Agarwal, James Balter, Yue Cao. Deep Learning based Joint MR Image Reconstruction and Under-sampling Pattern Optimization. Proceedings of ISMRM 2021.
- J. Zou, J. Balter, and Y. Cao. Joint Optimization of Subsampling Pattern and Reconstruction of DCE MRI for Pharmacokinetic Parameter Estimation. AAPM 2021 (Accepted).
Physiological Imaging Guided Radiation Therapy
Since 2003, under the leadership of Yue Cao, we have established a core team with expertise and infrastructure (software tool of imFIAT) to support in-house physiological imaging guided clinical trials with finical supports of NIH grants of P01 CA59827(Ten Haken), U01 CA183848 (Cao), R01 CA184153 (Cao), RO1 CA132834 (Cao) and RO1 NS064973 (Cao), and the Department of Radiation Oncology.
We have gone through multiple phases, from developing, testing and validating imaging techniques, to implementing and supporting the use of the techniques as imaging biomarkers in the clinical trials. The software tool, imFIAT, has been granted the level-5 benchmark by NIC Quantitative Image Network. We have supported image acquisition and analysis for clinical decision making in the trials, and completed three trials (heptocellular carcinoma (HCC, NCT02460835), locally advanced head-and-neck cancers (HNC, NCT02031250) and glioblastoma (GBM, NCT02805179)). Currently, we have several trials on-going in which imaging biomarkers are used as a decision making tool for adaptive radiation therapy (UMCC2017.113 (HNC), UMCC2020.033(GBM), and UMCC2020.052 (HCC)). We have automated the image processing procedures in imFIAT to support these trials and for the future trials.
Select Publications
Glioblastoma and Brain Function Responses to Radiation Therapy
- Y. Cao, C. I. Tsien, V. Nagesh, L. Junck, R. Ten Haken, B. D. Ross, T. L. Chenevert, and T. S. Lawrence. Survival prediction in high-grade gliomas by perfusion MRI prior to and during early stage of RT. Int J Rad Onc Biol Phys 64(3):876-885, 2006. PMID: 16298499
- Tong Zhu, Christopher H Chapman, Christina Tsien, Michelle Kim, Daniel E Spratt, Theodore S Lawrence, Yue Cao. Effect of the Maximum Dose on White Matter Fiber Bundles Utilizing Longitudinal Diffusion Tensor Imaging. Int J Rad Onbc Biol Phys, 96(3):696-705,2016. PMID: 27681767. PMCID: PMC5117804. DOI: 10.1016/j.ijrobp.2016.07.010
- Michelle M. Kim, Yilun Sun, Madhava P. Aryal, Hemant A. Parmar, Morand Piert, Benjamin Rosen, Charles S. Mayo, James M. Balter, Matthew Schipper, Nicolette Gabel, Emily Briceño, Daekeun You, Jason Heth, Wajd Al-Holou, Yoshie Umemura, Denise Leung, Larry Junck, Daniel R. Wahl, Theodore S. Lawrence, Yue Cao. A Phase II Study of Dose-Intensified Chemoradiation Using Biologically-Based Target Volume Definition in Patients with Newly Diagnosed Glioblastoma. Int J Rad Onc Biol Phys 2021. https://doi.org/10.1016/j.ijrobp.2021.01.033
- Michelle M. Kim, Madhava P. Aryal, Yilun Sun, Hemant A. Parmar, Pin Li, Matthew Schipper, Daniel R. Wahl, Theodore S. Lawrence, Yue Cao. Response assessment during chemoradiation using a hypercellular/hyperperfused advanced imaging phenotype predicts overall survival in patients with newly diagnosed glioblastoma. Neuro-Oncology 2021. https://doi.org/10.1093/neuonc/noab038
Hepatic Cancer and Liver Function Responses to Radiation Therapy
- Y. Cao, H. Wang, T.D. Johnson, C. Pan, H. Hussain, J.M. Balter, Daniel. Normolle, E. Ben-Josef, R. K. Ten Haken, T.S. Lawrence, and M. Feng. Prediction of Liver function using MR-based portal venous perfusion imaging. Int J Rad Onc Biol Phys 85(1):258-63, 2013. PMCID: PMC3629546. PMID: 22520476. DOI: 10.1371/journal.pone0057768
- H. Wang, R. Farjam, M. Feng, T. S. Lawrence, and Y. Cao. Arterial Perfusion Imaging-Defined Subvolume of Intrahepatic Cancer. Int J Rad Onbc Biol Phys, 89(1):167-174, 2014. PMCID: PMC3988211. PMID: 24613814. DOI: 10.1016/j.ijrobp.2014.01.040
- Josiah Simeth, Adam Johansson, Dawn Owen, Kyle Cuneo, Michelle Mierzwa, Mary Feng, Theodore S. Lawrence, and Yue Cao. Quantification of liver function by linearization of a 2-compartment model of gadoxetic-acid uptake using dynamic contrast enhanced magnetic resonance imaging. NMR in Biomedicine, 31(6), e3913, 2018.
Head and Neck Cancers and Normal Structures Responses to Radiation Therapy
- Y. Cao, A. Popovzter, D. Li, D.B. Chepeha, J.S. Moyer, M.E. Prince, F. Worden, T. Teknos, C. Bradford, S. K. Mukherji, and A. Eisbruch. Early prediction of outcome in advanced head and neck cancer by tumor blood volume alteration during radiation therapy. Int J Rad Onc Biol Phys 72(5):1287-1290, 2008. PMID: 19028268
- Yue Cao, Madhava Aryal, Pin Li, Choonik Lee, Matthew Schipper, Peter G. Hawkins, Christina Chapman, Dawn Owen, Aleksandar F. Dragovic, Paul Swiecicki, Keith Casper, Francis Worden, Lawrence S. Theodore, Avraham Eisbruch, Michelle Mierzwa. Predictive Values of MRI and PET Derived Quantitative Parameters for Patterns of Failure in both p16+ and p16- High Risk Head and Neck Cancer. Frontier in Oncology, 9:1118, 2019. http://doi.org/10.3389/fonc.2019.01118
- Michelle L Mierzwa, Laila A Gharzai, Pin Li, Joel Wilkie, Peter G Hawkins, Madhava P. Aryal, Choonik Lee, Teresa Lyden, Anna Blakely, Jennifer Tharamus, Matthew J Schipper, Christina H Chapman, Caitlin Schonewolf, Avraham Eisbruch, Yue Cao. Early blood volume changes in constrictor muscles correlate with post-radiation dysphagia. Int J Rad Onc Biol Phys 2021 (in press).
Development of MR-based in vivo Microstructural Imaging
MR-based in vivo microstructural imaging is an emerging technology for quantification of tissue microstructure and restriction effects of water diffusion, but to date has limited development on clinical scanner hardware and application in patients with tumors. We aim to develop, evaluate, and validate in vivo microstructural imaging, including acquisition and modeling. We have been optimizing diffusion signal acquisition parameters of b-value, diffusion time and TE using a prototype pulse sequence with oscillating diffusion gradient capability. We investigate computation models to estimate the microstructural and diffusion parameters in brain, head-and-neck and liver tumors. We are interested in the parameters, including cell size (ls ≈ const x V/S (a ratio of volume to surface)), cell membrane permeability (κ), free diffusion coefficient (D0), bulk diffusion coefficient (Dinf), and effective restriction (ξ). We initiate a project to validate the estimated microstructural parameters from in vivo imaging with ones from digital pathology analysis of resected tissue samples through collaborations with surgeons and pathologists. We will further evaluate whether these microstructural parameters can be used as imaging biomarkers for cancer therapy assessment.
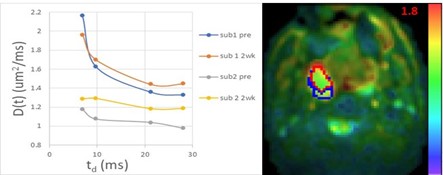
Left: Time-dependent diffusion coefficients with diffusion times of 6.9-28.1 ms in two subregions of a patient with head-and neck cancer (HNC) pre-radiation therapy (RT) and at week 2 during RT. Right: An example of Dinf maps estimated using a long-time limit of the random walk with barriers model. Blue and red contours depict 2 subregions of a tumor (sub1: blue; sub2: red).

Dr. Balis’ lab is capable of performing pixel level classification at massively-parallel spatial computational scale to extract specific biological contextual features. (A-B) Representative fields of view from overall 25 mm2 sections of tissue of benign (A) and malignant (B) regions of an oral squamous cell carcinoma. Cellular cross-sectional area distributions are plotted from both fields of view and show benign (blue) and malignant (organ) peaks at 60 and 300 um2, respectively, which can be used to validate in vivo estimations.
Members:
Group Members
- Yue Cao, PhD Professor
- James Balter, PhDProfessor
- Hesheng Wang, PhD, Research Investigator
- Antonis Matakos, PhDPost-doctoral research fellow
- Priyanka Pramanik, MS Image Analyst
- Lianli Liu, BS Grad Student Research Asst.
- Ross Avila, BS, Medical School Student
New Members

- Deakeung You, PhD, Post-doctoral research fellow in Sept. 2014
- Madhava P Aryal, PhD, Post-doctoral research fellow in Sept. 2014
- Tong Zhu, PhD, Post-doctoral research fellow (Med Phys Resident) in Sept. 2014
- Eric Paradis, PhD, Post-doctoral research fellow in Oct. 2014
- Adam Johansson, PhD, Candidate, Post-doctoral research fellow in Jan. 2015
Alumni
- Vijaya Nagesh, PhD, Junior faculty
- P Wang, PhD, Post-doctoral/physics resident fellow
- Chris Chapman, MD, Research assistant
- Jonathan Alspaugh, MS, Undergraduate and post-graduate research assistant
- Diana Li, MS, Undergraduate and post-graduate research assistant
- John Hayes, Undergraduate research assistant
- Kyle McMillan, Undergraduate research assistant
- Orit Gutfeld, MD, Visiting clinical fellow
- Ellen Kerkhof, Visiting doctoral student from the University of Utrecht, The Netherlands
- Reza Farjam, PhD, Student of BME and post-doctoral research fellow
- Colleen Fox, PhD, Post-doctoral/physics resident fellow
- Mohammad Nazemzaseh, PhD, Post-doctoral research fellow
- Ke Huang, PhD, Post-doctoral/physics resident fellow
- Krithika Shanmugasundaram, Medical school student research assistant
- Shu-Hui Hsu, PhD, Post-doctoral research fellow
- Chris Chapman, MD, medical school student
Join Us:
Laboratory Lead

