Abstract
XX female and XY male therian mammals equalize X-linked gene expression through the mitotically-stable transcriptional inactivation of one of the two X chromosomes in female somatic cells. Here, we describe an essential function of the X-linked homolog of an ancestral X-Y gene pair, Kdm5c-Kdm5d, in the expression of Xist lncRNA, which is required for stable X-inactivation. Ablation of Kdm5c function in females results in a significant reduction in Xist RNA expression. Kdm5c encodes a demethylase that enhances Xist expression by converting histone H3K4me2/3 modifications into H3K4me1. Ectopic expression of mouse and human KDM5C, but not the Y-linked homolog KDM5D, induces Xist in male mouse embryonic stem cells (mESCs). Similarly, marsupial (opossum) Kdm5c but not Kdm5d also upregulates Xist in male mESCs, despite marsupials lacking Xist, suggesting that the KDM5C function that activates Xist in eutherians is strongly conserved and predates the divergence of eutherian and metatherian mammals. In support, prototherian (platypus) Kdm5c also induces Xist in male mESCs. Together, our data suggest that eutherian mammals co-opted the ancestral demethylase KDM5C during sex chromosome evolution to upregulate Xist for the femalespecific induction of X-inactivation.
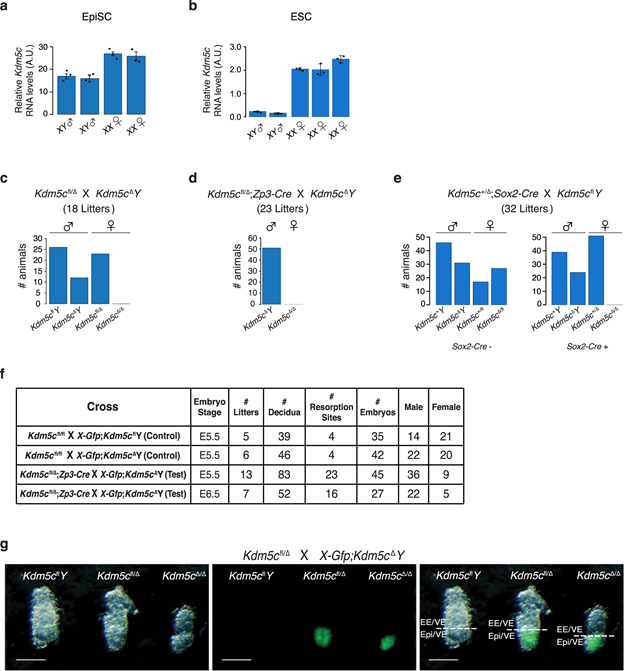
Fig. 1: Lethality of Kdm5cΔ/Δ female embryos.
Read the full text of this article here - Activation of Xist by an evolutionarily conserved function of KDM5C demethylase
Samanta, M.K., Gayen, S., Harris, C. et al. Activation of Xist by an evolutionarily conserved function of KDM5C demethylase. Nat Commun 13, 2602 (2022). https://doi.org/10.1038/s41467-022-30352-1
Milan Kumar Samanta 1,6, Srimonta Gayen 1,4,6, Clair Harris1, Emily Maclary1,5, Yumie Murata-Nakamura1, Rebecca M. Malcore1, Robert S. Porter 1, Patricia M. Garay 1,2, Christina N. Vallianatos 1, Paul B. Samollow3, Shigeki Iwase 1,2 & Sundeep Kalantry 1 ✉
1 Department of Human Genetics, University of Michigan Medical School, Ann Arbor, MI 48109-5618, USA. 2 Neuroscience Graduate Program, University of Michigan Medical School, Ann Arbor, MI 48109-5618, USA. 3 Department of Veterinary Integrative Biosciences, College of Veterinary Medicine and Biomedical Sciences, Texas A&M University, College Station, TX 77843-4458, USA. 4Present address: Department of Molecular Reproduction, Development and Genetics, Indian Institute of Science, Bangalore, Karnataka 560012, India. 5Present address: Department of Biology, University of Utah, Salt Lake City, UT 84112, USA. 6These authors contributed equally: Milan Kumar Samanta, Srimonta Gayen. ✉email: [email protected]


Shigeki Iwase, Ph.D.

