The group found four patient missense variants at adjacent codons in the key pituitary TF POU1F1, a leading genetic cause of hypopituitarism. These turned out to shift splicing to favor the minor beta isoform, a transcriptional repressor.
We wondered how widespread this effect was, and developed a splicing MAVE to measure how each of the >1000 possible SNVs in and around this exon affected splicing.
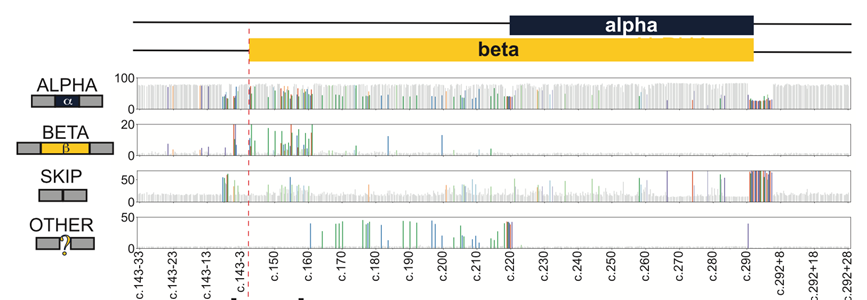
Map in hand, we revisited patient sequencing results, and found two affected individuals carrying ultra-rare, splice-disruptive synonymous variants. In total we find ~9% the tested SNVs are splice disruptive, and nearly 1 in 7 of these are synonymous.
We hope this will be a useful resource, both for resolving future POU1F1 VUSs, and also for developing improved wet- and dry-lab methods for splice effect prediction. Congrats to everyone involved!
